Comparative Molecular Transporter Properties of Cyclic Peptides Containing Tryptophan and Arginine Residues Formed through Disulfide Cyclization
Abstract
:1. Introduction
2. Results and Discussion
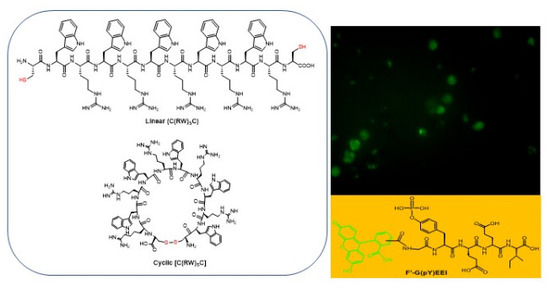
2.1. Chemistry
2.2. In Vitro Cytotoxicity Assay of Peptides
2.3. Cellular Uptake Studies
2.3.1. Cellular Uptake of Fluorescent-Labeled Compounds in the Presence of Synthesized Peptides
2.3.2. Fluorescence Microscopy
3. Conclusions
4. Materials and Methods
4.1. Materials
4.2. Chemistry
4.2.1. Synthesis of Linear Peptides (C(WR)4C) and (C(WR)5C)
4.2.2. Synthesis of Cyclic Peptides ([C(WR)4C], [C(WR)5C])
4.3. In Vitro Cytotoxicity Assay of Peptides
4.4. Cellular Uptake Studies
4.4.1. Mechanism of Cellular Uptake: Effect of Endocytic Inhibitors
4.4.2. Fluorescence Microscopy
4.5. Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Zhou, Y.Y.; Abagyan, R. How and why phosphotyrosine-containing peptides bind to the SH2 and PTB domains. Fold. Des. 1998, 3, 513–522. [Google Scholar] [CrossRef] [Green Version]
- Songyang, Z.; Shoelson, S.E.; Chaudhuri, M.; Gish, G.; Pawson, T.; Haser, W.G.; King, F.; Roberts, T.; Ratnofsky, S.; Lechleider, R.J.; et al. SH2 domains recognize specific phosphopeptide sequences. Cell 1993, 72, 767–778. [Google Scholar] [CrossRef] [PubMed]
- Tinti, M.; Kiemer, L.; Costa, S.; Miller, M.L.; Sacco, F.; Olsen, J.V.; Carducci, M.; Paoluzi, S.; Langone, F.; Workman, C.T.; et al. The SH2 domain interaction landscape. Cell Rep. 2013, 3, 1293–1305. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mukherjee, S.; Ray, S.; Thakur, R.S. Solid lipid nanoparticles: A modern formulation approach in drug delivery system. Indian J. Pharm. Sci. 2009, 71, 349–358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deepa, K.; Singha, S.; Panda, T. Doxorubicin nanoconjugates. J. Nanosci. Nanotechnol. 2014, 14, 892–904. [Google Scholar] [CrossRef] [PubMed]
- Vasconcelos, L.; Parn, K.; Langel, U. Therapeutic potential of cell-penetrating peptides. Ther. Deliv. 2013, 4, 573–591. [Google Scholar] [CrossRef]
- Derakhshankhah, H.; Jafari, S. Cell penetrating peptides: A concise review with emphasis on biomedical applications. Biomed. Pharmacother. 2018, 108, 1090–1096. [Google Scholar] [CrossRef]
- Guidotti, G.; Brambilla, L.; Rossi, D. Cell-Penetrating Peptides: From Basic Research to Clinics. Trends Pharmacol. Sci. 2017, 38, 406–424. [Google Scholar] [CrossRef]
- Kardani, K.; Milani, A.; Shabani, S.H.; Bolhassani, A. Cell penetrating peptides: The potent multi-cargo intracellular carriers. Expert Opin. Drug Deliv. 2019, 16, 1227–1258. [Google Scholar] [CrossRef]
- Pujals, S.; Fernandez-Carneado, J.; Lopez-Iglesias, C.; Kogan, M.J.; Giralt, E. Mechanistic aspects of CPP-mediated intracellular drug delivery: Relevance of CPP self-assembly. Biochim. Biophys. Acta 2006, 1758, 264–279. [Google Scholar] [CrossRef] [Green Version]
- Pooga, M.; Langel, U. Classes of Cell-Penetrating Peptides. Methods Mol. Biol. 2015, 1324, 3–28. [Google Scholar] [CrossRef] [PubMed]
- Rothbard, J.B.; Jessop, T.C.; Lewis, R.S.; Murray, B.A.; Wender, P.A. Role of membrane potential and hydrogen bonding in the mechanism of translocation of guanidinium-rich peptides into cells. J. Am. Chem. Soc. 2004, 126, 9506–9507. [Google Scholar] [CrossRef] [PubMed]
- Rydberg, H.A.; Matson, M.; Amand, H.L.; Esbjorner, E.K.; Norden, B. Effects of Tryptophan Content and Backbone Spacing on the Uptake Efficiency of Cell-Penetrating Peptides. Biochemistry-Us 2012, 51, 5531–5539. [Google Scholar] [CrossRef] [PubMed]
- Favretto, M.E.; Wallbrecher, R.; Schmidt, S.; van de Putte, R.; Brock, R. Glycosaminoglycans in the cellular uptake of drug delivery vectors—Bystanders or active players? J. Control. Release 2014, 180, 81–90. [Google Scholar] [CrossRef] [PubMed]
- Raucher, D.; Ryu, J.S. Cell-penetrating peptides: Strategies for anticancer treatment. Trends Mol. Med. 2015, 21, 560–570. [Google Scholar] [CrossRef]
- Conner, S.D.; Schmid, S.L. Regulated portals of entry into the cell. Nature 2003, 422, 37–44. [Google Scholar] [CrossRef]
- Deshayes, S.; Plenat, T.; Aldrian-Herrada, G.; Divita, G.; Le Grimellec, C.; Heitz, F. Primary amphipathic cell-penetrating peptides: Structural requirements and interactions with model membranes. Biochemistry-Us 2004, 43, 7698–7706. [Google Scholar] [CrossRef]
- Nakase, I.; Niwa, M.; Takeuchi, T.; Sonomura, K.; Kawabata, N.; Koike, Y.; Takehashi, M.; Tanaka, S.; Ueda, K.; Simpson, J.C.; et al. Cellular uptake of arginine-rich peptides: Roles for macropinocytosis and actin rearrangement. Mol. Ther. 2004, 10, 1011–1022. [Google Scholar] [CrossRef]
- Verdurmen, W.P.; Bovee-Geurts, P.H.; Wadhwani, P.; Ulrich, A.S.; Hallbrink, M.; van Kuppevelt, T.H.; Brock, R. Preferential uptake of L- versus D-amino acid cell-penetrating peptides in a cell type-dependent manner. Chem. Biol. 2011, 18, 1000–1010. [Google Scholar] [CrossRef] [Green Version]
- Walrant, A.; Bauza, A.; Girardet, C.; Alves, I.D.; Lecomte, S.; Illien, F.; Cardon, S.; Chaianantakul, N.; Pallerla, M.; Burlina, F.; et al. Ionpair-pi interactions favor cell penetration of arginine/tryptophan-rich cell-penetrating peptides. Biochim. Biophys. Acta Biomembr. 2020, 1862, 183098. [Google Scholar] [CrossRef]
- Patil, K.M.; Naik, R.J.; Vij, M.; Yadav, A.K.; Kumar, V.A.; Ganguli, M.; Fernandes, M. Second generation, arginine-rich (R-X’-R)(4)-type cell-penetrating alpha-omega-alpha-peptides with constrained, chiral omega-amino acids (X’) for enhanced cargo delivery into cells. Bioorg. Med. Chem. Lett. 2014, 24, 4198–4202. [Google Scholar] [CrossRef] [PubMed]
- Dougherty, P.G.; Sahni, A.; Pei, D. Understanding Cell Penetration of Cyclic Peptides. Chem. Rev. 2019, 119, 10241–10287. [Google Scholar] [CrossRef] [PubMed]
- Park, S.E.; Sajid, M.I.; Parang, K.; Tiwari, R.K. Cyclic Cell-Penetrating Peptides as Efficient Intracellular Drug Delivery Tools. Mol. Pharm. 2019, 16, 3727–3743. [Google Scholar] [CrossRef] [PubMed]
- Mandal, D.; Nasrolahi Shirazi, A.; Parang, K. Cell-penetrating homochiral cyclic peptides as nuclear-targeting molecular transporters. Angew. Chem. Int. Ed. Engl. 2011, 50, 9633–9637. [Google Scholar] [CrossRef]
- Shirazi, A.N.; Tiwari, R.K.; Oh, D.; Banerjee, A.; Yadav, A.; Parang, K. Efficient Delivery of Cell Impermeable Phosphopeptides by a Cyclic Peptide Amphiphile Containing Tryptophan and Arginine. Mol. Pharmaceut. 2013, 10, 2008–2020. [Google Scholar] [CrossRef] [Green Version]
- Shirazi, A.N.; Tiwari, R.; Chhikara, B.S.; Mandal, D.; Parang, K. Design and Biological Evaluation of Cell-Penetrating Peptide-Doxorubicin Conjugates as Prodrugs. Mol. Pharmaceut. 2013, 10, 488–499. [Google Scholar] [CrossRef]
- Shirazi, A.N.; Tiwari, R.K.; Oh, D.; Sullivan, B.; Kumar, A.; Beni, Y.A.; Parang, K. Cyclic Peptide-Selenium Nanoparticles as Drug Transporters. Mol. Pharmaceut. 2014, 11, 3631–3641. [Google Scholar] [CrossRef]
- Mozaffari, S.; Bousoik, E.; Amirrad, F.; Lamboy, R.; Coyle, M.; Hall, R.; Alasmari, A.; Mahdipoor, P.; Parang, K.; Aliabadi, H.M. Amphiphilic Peptides for Efficient siRNA Delivery. Polymers 2019, 11, 703. [Google Scholar] [CrossRef] [Green Version]
- Bansal, A.; Simon, M.C. Glutathione metabolism in cancer progression and treatment resistance. J. Cell Biol. 2018, 217, 2291–2298. [Google Scholar] [CrossRef] [Green Version]
- Traverso, N.; Ricciarelli, R.; Nitti, M.; Marengo, B.; Furfaro, A.L.; Pronzato, M.A.; Marinari, U.M.; Domenicotti, C. Role of glutathione in cancer progression and chemoresistance. Oxid. Med. Cell Longev. 2013, 2013, 972913. [Google Scholar] [CrossRef] [Green Version]
- Machida, K.; Mayer, B.J. The SH2 domain: Versatile signaling module and pharmaceutical target. Biochim. Biophys. Acta 2005, 1747, 1–25. [Google Scholar] [CrossRef]
- Agarwal, H.K.; Loethan, K.; Mandal, D.; Doncel, G.F.; Parang, K. Synthesis and biological evaluation of fatty acyl ester derivatives of 2 ’,3 ’-didehydro-2 ’,3 ’-dideoxythymidine. Bioorg. Med. Chem. Lett. 2011, 21, 1917–1921. [Google Scholar] [CrossRef]
- Agarwal, H.K.; Chhikara, B.S.; Bhavaraju, S.; Mandal, D.; Doncel, G.F.; Parang, K. Emtricitabine Prodrugs with Improved Anti-HIV Activity and Cellular Uptake. Mol. Pharmaceut. 2013, 10, 467–476. [Google Scholar] [CrossRef]
- Jones, S.W.; Christison, R.; Bundell, K.; Voyce, C.J.; Brockbank, S.M.V.; Newham, P.; Lindsay, M.A. Characterisation of cell-penetrating peptide-mediated peptide delivery. Brit. J. Pharmacol. 2005, 145, 1093–1102. [Google Scholar] [CrossRef]
- Thorn, C.F.; Oshiro, C.; Marsh, S.; Hernandez-Boussard, T.; McLeod, H.; Klein, T.E.; Altman, R.B. Doxorubicin pathways: Pharmacodynamics and adverse effects. Pharmacogenet. Genom. 2011, 21, 440–446. [Google Scholar] [CrossRef]
- Agarwal, H.K.; Chhikara, B.S.; Hanley, M.J.; Ye, G.; Doncel, G.F.; Parang, K. Synthesis and biological evaluation of fatty acyl ester derivatives of (-)-2’,3’-dideoxy-3’-thiacytidine. J. Med. Chem. 2012, 55, 4861–4871. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds are available from the authors depending on the availability. |












© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mohammed, E.H.M.; Mandal, D.; Mozaffari, S.; Abdel-Hamied Zahran, M.; Mostafa Osman, A.; Kumar Tiwari, R.; Parang, K. Comparative Molecular Transporter Properties of Cyclic Peptides Containing Tryptophan and Arginine Residues Formed through Disulfide Cyclization. Molecules 2020, 25, 2581. https://doi.org/10.3390/molecules25112581
Mohammed EHM, Mandal D, Mozaffari S, Abdel-Hamied Zahran M, Mostafa Osman A, Kumar Tiwari R, Parang K. Comparative Molecular Transporter Properties of Cyclic Peptides Containing Tryptophan and Arginine Residues Formed through Disulfide Cyclization. Molecules. 2020; 25(11):2581. https://doi.org/10.3390/molecules25112581
Chicago/Turabian StyleMohammed, Eman H. M., Dindyal Mandal, Saghar Mozaffari, Magdy Abdel-Hamied Zahran, Amany Mostafa Osman, Rakesh Kumar Tiwari, and Keykavous Parang. 2020. "Comparative Molecular Transporter Properties of Cyclic Peptides Containing Tryptophan and Arginine Residues Formed through Disulfide Cyclization" Molecules 25, no. 11: 2581. https://doi.org/10.3390/molecules25112581